In Canada, around 300 million spruces are planted annually, representing over 100,000 hectares reforested annually. Spruce planting has remained unimproved for complex traits such as wood quality or resilience to biotic and climatic factors, because of the high assessment costs and long evaluation time in field testing. With the progress made in spruce genomics, the timing is right for major forest sector players to partner in moving genomic selection tests (GS-tests) toward application at an operational scale. GS-tests makes it possible to predict future performance of young trees and thus speed up selection, ensuring new plantation stock is more productive and resilient in the context of climate change.
Genomic selection tests (GS-tests)—in this project called FastTRAC for Fast Tests for Rating and Amelioration of Conifers (in French, Tests Rapides pour l’Amélioration des Conifères) — are diagnostic tools/procedures that were developed in Quebec over the last 10 years for spruces, and make it possible to rapidly predict the genetic value of trees used in breeding programs at a very young age and for very large number of trees and for a variety of traits simultaneously.
The procedure is based on multi-loci genotypic information called genomic profiles (see diagram). The FastTRAC approach differs from conventional breeding in that an estimation of trees’ genetic value can be obtained at a very young age (or seedling stage) through their genomic profiles, which can be obtained in a few months, instead of waiting for them being evaluated in the field for up to 30 years for some economically and environmentally important traits.
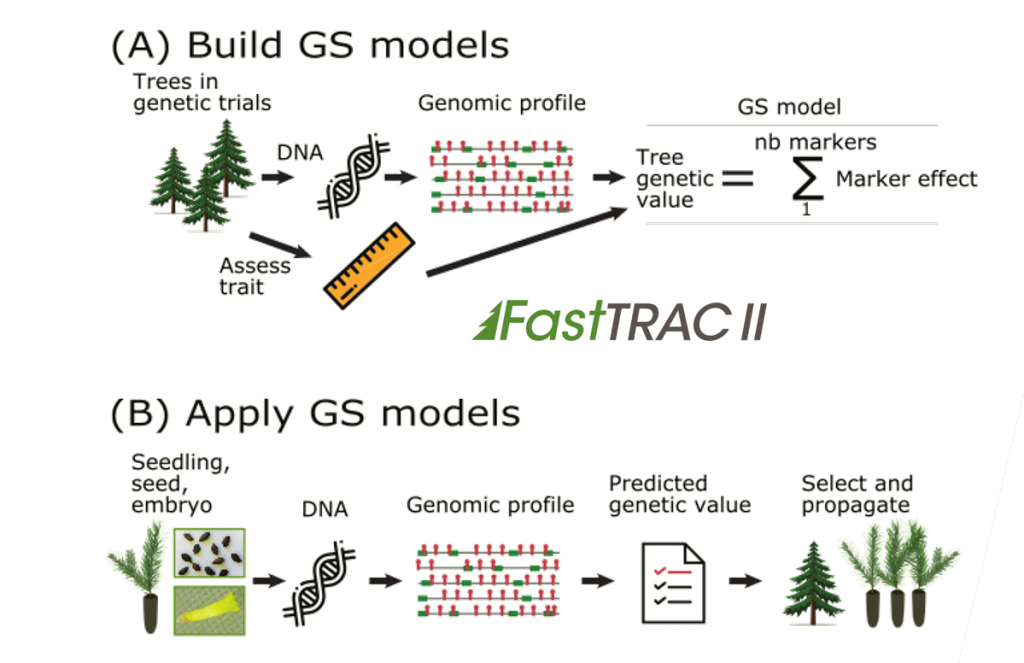
To develop for a species and a breeding population a GS-test for a trait of economic or adaptive importance such as growth productivity, wood density or resilience to biotic or climatic factors, the trait must be assessed from few hundred mature trees present in plantations established by tree breeders in the past. Next, the unique genomic profiles of trees for which the trait of interest was assessed must be obtained using a large number of genetic markers.
A mathematical model is then built to enable the genomic profiles of trees to be “linked” with their plantation trait values. After validation using independent data from the same mature plantation, the GS-test can then be used to predict the genetic value of any other tree from the same population, the latter being at any stage (such as young seedlings or embryogenic cell lines). To carry out the GS-test, only a small amount of DNA of each candidate tree is needed from which its genomic profile will be obtained.
Results of our recent studies have shown that from 90% to 100% of the gain normally obtained with the conventional rating approach using field testing over long periods of time (tens of years) can be obtained with genomic FastTRACs applied to young trees or seedlings of the same breeding population without having to evaluate them in the field. Young candidate trees for which the genetic value has been predicted using their genomic profiles can be ranked relative to each other and the best ones can be selected based one trait or more, and then be vegetatively reproduced and used in reforestation programs without having to wait for their mature stage assessment. Moreover, success of vegetative propagation is much higher with young trees.
Genomic selection makes it possible to generate very significant economic gains because plantations with new genetically improved stock can take place one generation earlier while giving more flexibility to tree breeders to tailor their improved genetic stock as climate change evolves, or to develop new breeding objectives in order to meet demand for new end-products on global markets.